Research
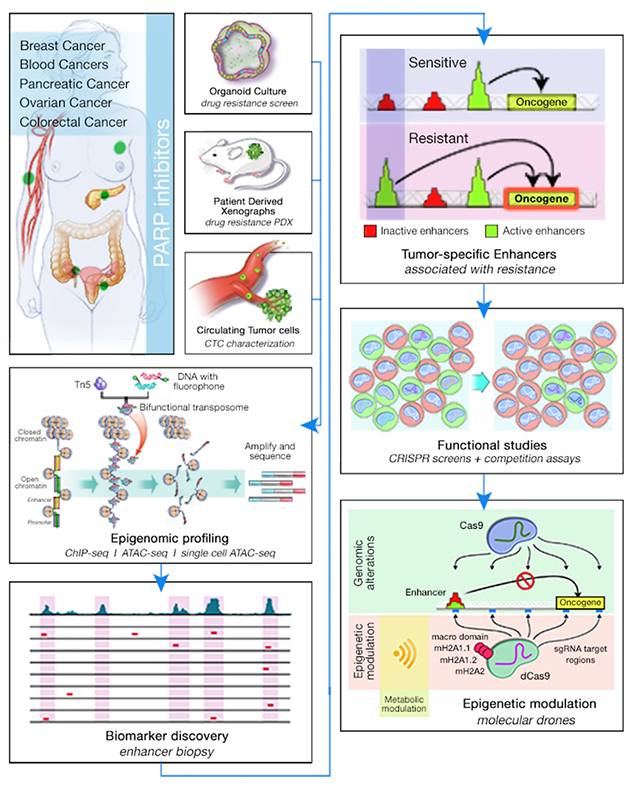
Along with genetic tests, epigenomic profiling has become one of the most important areas in the advancing efforts for personalized medicine. Novel epigenomic techniques have led to a better understanding of how expression patterns are regulated by epigenetic changes from tissues to organs and across people and health states. These expression changes are, in part, mediated by specialized cis-regulatory elements called enhancers, which contain DNA motifs recognized by transcription factors to activate transcription.
Enhancer modulation
Epigenomic mapping has revealed unexpectedly complex and dynamic enhancer patterns that share certain stereotypical chromatin features, such as open chromatin, and enrichment for the regulatory histone markers H3K4me1 and H3K27ac (for active enhancers). Epigenomic mapping allows for the identification and characterization of transcriptional dependencies in cancer that could potentially be targeted with therapies.
The Functional Epigenomics Lab has been working on various aspects of transcription and enhancer regulation with implications in cellular heterogeneity, malignant transformation and reprogramming. The goal is to use epigenomic profiling to better understand cancer programs associated with malignancy, metastasis and drug sensitivity, and define transcriptional dependencies.
To achieve this long-term goal, the lab is using three complementary avenues:
- Mechanistic studies. Using CRISPR/Cas9-based technologies and coculturing systems to address cellular heterogeneity, the lab aims to target novel transcription factor candidates and noncoding elements of the genome to functionally validate their roles in drug sensitivity. Coupling time-lapse culturing technologies with advanced culturing systems using microfluidic devices allows the lab to test different drug combinations.
- Technology development. By adapting the most recent advances in sequencing technologies to a variety of model systems — 3D organoids, patient-derived xenografts and liquid biopsy — the lab aims to address tumor heterogeneity and epigenomic profiling in small populations of cells.
- Bioinformatic analysis. With a special interest in incorporating different sequencing platforms to identify epigenomic patterns, the lab focuses on extracting the most information from RNA-seq, ChIP-seq, HiC, ATAC-seq and single-cell ATAC-seq-RNA-seq data. One important focus of the lab's analysis is the integration of cell-specific datasets from publicly available data in the lab's single-cell analysis to better define regulatory mechanisms that are only relevant for specific cell types.