-

The CosMx Spatial Molecular Imaging platform defines the boundaries of cells in a triple-negative breast cancer sample. This process is called segmentation. Learn about the facility's services.
-
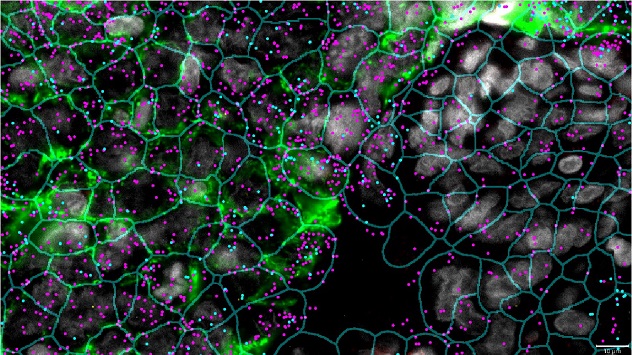
Two major histocompatibility complex transcripts are overlaid on the cell segmentation pattern: magenta for the HLA-A gene and blue for the HLA-DRA gene. Learn about our equipment.
-

Each cell was identified by studying transcript profiles: tumor cells, red; macrophages, green; endothelial cells, brown; and fibroblasts, gray. See the facility's faculty.
Overview
The Spatial Biology Core evolved from studying the relationship between how genes function during the process of transformation of normal cells to cancer cells. The facility offers investigators tools and collaboration for conducting basic cellular research. This testing enables a better understanding of cancer cells and reveals possibilities for developing novel treatments.
The facility was one of the first to use NanoString Technologies' nCounter barcoding technology. This technology uses color-coded molecular barcodes that can hybridize to many types of target molecules.
Over the years, the facility has done alpha and beta testing of most of the cancer-related products created and marketed by NanoString Technologies. One of the facility's major services is analyzing messenger RNA abundance in formalin-fixed, paraffin-embedded (FFPE) clinical samples.
The Spatial Biology Core has long had a productive collaboration with NanoString Technologies. The facility was one of four sites to beta-test the GeoMx Digital Spatial Profiling platform. Beta testing began in September 2019 and led to the first commercial instrument shipping in January 2020.
The facility has since used the GeoMx platform to process samples from more than 4,000 people. Most are FFPE samples from solid tumors, including breast, lung, colon, pancreatic, hepatocellular and ovarian cancers and cholangiocarcinoma. During processing, the facility measures spatial expression of protein and RNA. Spatial context can refer to the 2D or 3D space within tissue. This helps researchers understand the complex interactions between cells and molecularly characterize processes, cells and genes.
The facility has studied:
- Full-face tumor sections, as well as diagnostic core biopsies and tissue microarray samples.
- Routine and precancerous samples.
- The immune landscape or network of immune cells in neurodegenerative diseases of aging, using clinical samples and mouse models.
The facility works closely with its collaborators to make sure that these costly and time-consuming analyses are done in the best way possible. As many as 20 projects are underway or being planned at any time.
The Spatial Biology Core is working with NanoString Technologies to develop and deploy the CosMx Spatial Molecular Imaging platform. Using a prototype of this platform, the facility carried out an early-stage analysis of triple-negative breast cancer.
The facility continues to work with NanoString Technologies to make the plans of study that support the CosMx Spatial Molecular Imaging platform the best they can be. The facility works with NanoString Technologies collaborators to create and test analytic applications and data processing solutions. The facility was involved in beta-testing the CosMx instrument and working to put in place the cloud-based AtoMx software to study CosMx data.
Two CosMx instruments are in operation, measuring both RNA and protein at the single cell level with spatial context.